DOMINO is a domain-aware model calibration method that leverages semantic confusability and hierarchical similarity between class labels. In head MRI segmentation, DOMINO-calibrated networks outperform non-calibrated models and state-of-the-art morphometric methods, delivering better calibration, higher accuracy, and faster inference—especially on rarer classes. The performance stems from domain-aware regularization that informs semantic model calibration, improving trustworthiness and reliability of medical image segmentation models.
- Paper (arXiv): https://arxiv.org/abs/2209.06077
- Base model (MONAI UNETR): https://github.com/Project-MONAI/research-contributions/tree/main/UNETR
- DOMINO CLI: https://github.com/lab-smile/domino-cli
- Demo video: https://youtu.be/mKeXWM--xyU
- Code Ocean capsule: https://codeocean.com/capsule/6022409/tree/v2
- Request Pretrained Models: https://forms.gle/3GPnXXvWgaM6RZvr5
This repository provides the official implementation for training and using DOMINO from:
- DOMINO: Domain-aware Model Calibration in Medical Image Segmentation Skylar E. Stolte1, Kyle Volle2, Aprinda Indahlastari3,4, Alejandro Albizu3,5, Adam J. Woods3,4,5, Kevin Brink6, Matthew Hale2, and Ruogu Fang1,3,7* 1 J. Crayton Pruitt Family Department of Biomedical Engineering, Herbert Wertheim College of Engineering, University of Florida (UF), USA 2 Department of Mechanical and Aerospace Engineering, Herbert Wertheim College of Engineering, UF, USA 3 Center for Cognitive Aging and Memory, McKnight Brain Institute, UF, USA 4 Department of Clinical and Health Psychology, College of Public Health and Health Professions, UF, USA 5 Department of Neuroscience, College of Medicine, UF, USA 6 United States Air Force Research Laboratory, Eglin Air Force Base, Florida, USA 7 Department of Electrical and Computer Engineering, Herbert Wertheim College of Engineering, UF, USA MICCAI 2022
- DOMINO improves calibration and accuracy in head segmentation from T1 MRIs.
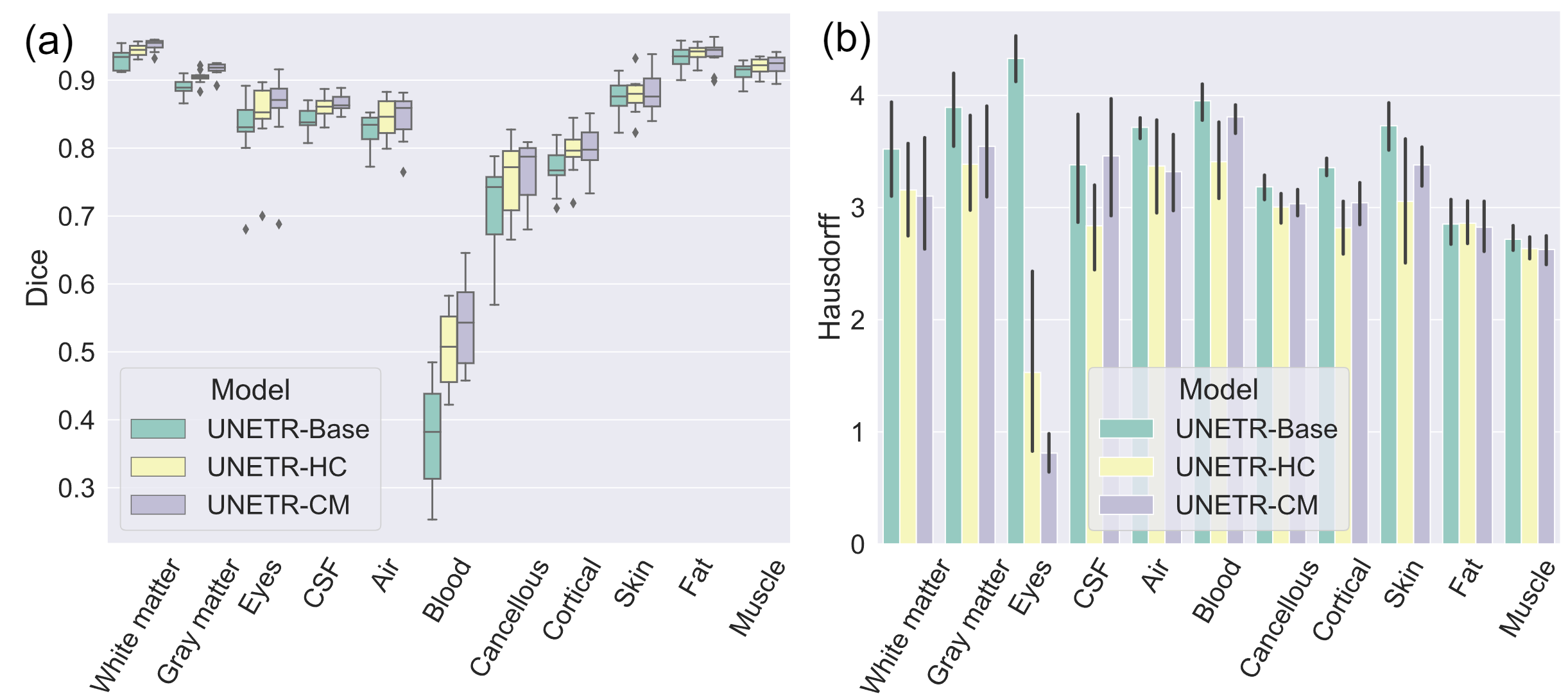
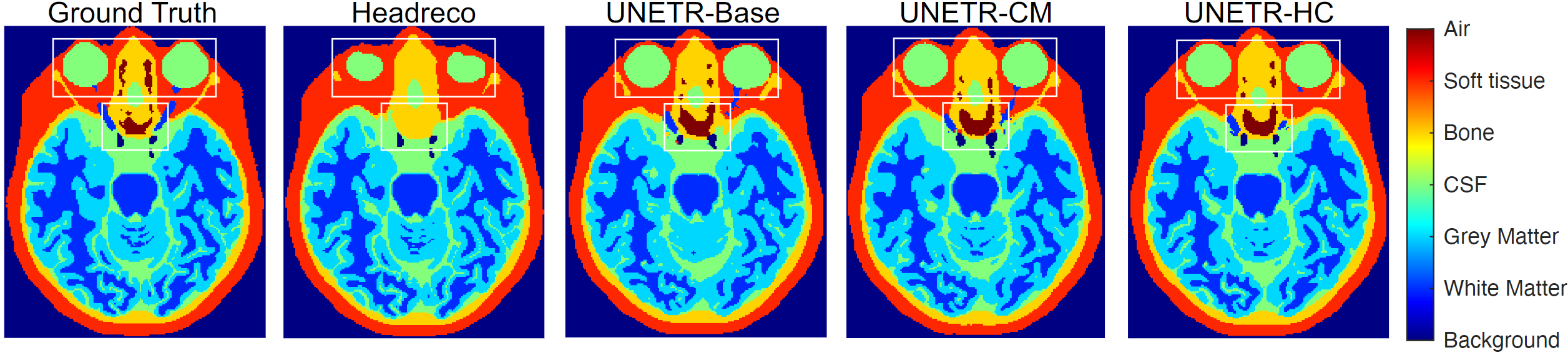
- DOMINO-CM: higher Top-1/Top-2/Top-3 accuracy than DOMINO-HC and an uncalibrated model (better regional performance and awareness of non-selected classes).
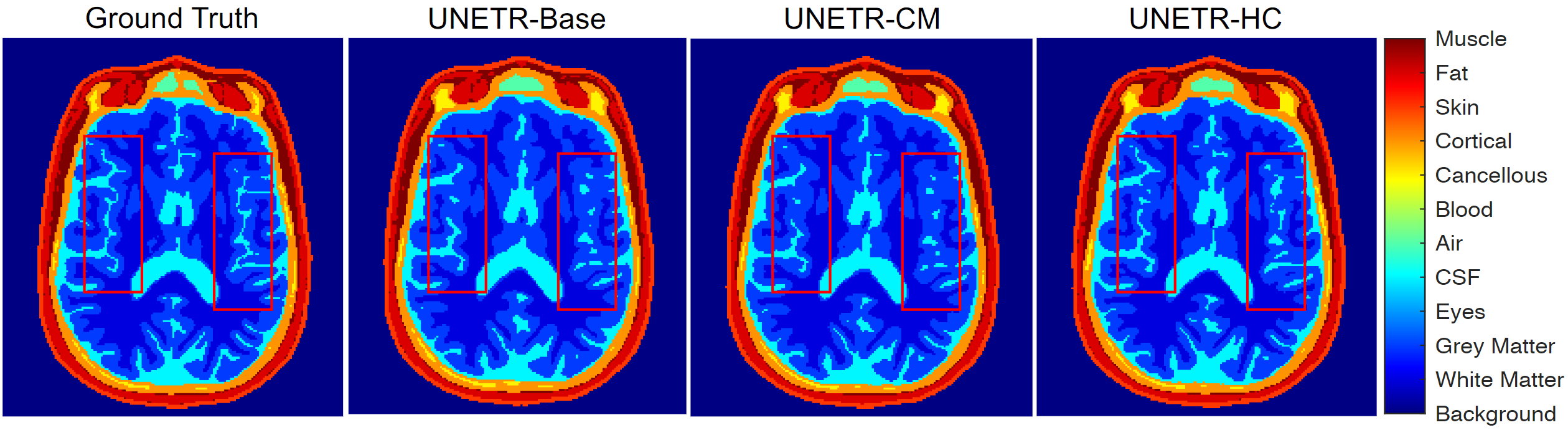
- DOMINO-HC: more precise boundary detection than DOMINO-CM and an uncalibrated model (critical where uncertainty is highest).
| Method | Top‑1 | Top‑2 | Top‑3 |
|---|---|---|---|
| HEADRECO | 0.905 | 0.977 | 0.983 |
| UNETR‑Base | 0.913 | 0.993 | 0.998 |
| UNETR‑HC | 0.924 | 0.995 | 0.998 |
| UNETR‑CM | 0.928 | 0.996 | 0.999 |
Figure 1: Top‑N accuracy on 6 classes.
Figure 2: (a) Dice scores and (b) Hausdorff distances in 11-class segmentation.
Figure 3: Sample image slice for 11-tissue segmentation.
Figure 4: Sample image slice for 6-tissue segmentation.
Two MATLAB scripts are included. Set the DOMINO working folder, add it to the MATLAB path, then:
- For MATLAB 2020b, change line 56 to:
image(index) = tissue_cond_updated.Labels(k)Run combine_mask.m. Expected output structure:
Data
ImagesTr sub-TrX_T1.nii, sub-TrXX_T1.nii, ...
ImagesTs sub-TsX_T1.nii, sub-TsXX_T1.nii, ...
LabelsTr sub-TrX_seg.nii, sub-TrXX_seg.nii, ...
LabelsTs sub-TsX_seg.nii, sub-TsXX_seg.nii, ...
Navigate to /your_data/Data/ and run make_datalist_json.m. Then exit MATLAB and proceed via terminal.
Following structure is required to run DOMINO, you may use preprocess.py for this. The preprocess.py script converts DOMINO-style raw data into nnU-Net format. Data directory must contain one or more source folders, each with subject folders named sub-, each containing T1 and mask files:
/path/to/your/data/ <-- --data_dir
├── source_folder_A/ <-- --source-folders
│ ├── sub-10001/
│ │ ├── T1.nii
│ │ └── T1_T1orT2_masks.nii
│ ├── sub-10002/
│ │ ├── T1.nii
│ │ └── T1_T1orT2_masks.nii
│ └── ...
└── source_folder_B/
├── sub-30001/
│ ├── T1.nii
│ └── T1_T1orT2_masks.nii
└── ...
- File Consolidation: Scans specified --source-folders for T1.nii and T1_T1orT2_masks.nii pairs, copies to temporary images/ and labels/ directories, and renames files as .nii (e.g., 10001.nii).
- Train-Test Split:
- Group A: subject IDs starting with “1” or “2”
- Group B: subject IDs starting with “3”
- Within each group: 90% train (Tr), 10% test (Ts) Creates nnU-Net folders: imagesTr, labelsTr, imagesTs, labelsTs.
- JSON Dataset Generation: Creates dataset.json; reserves 10% of training for validation, producing training, validation, and test splits in JSON.
Example:
singularity exec --nv --bind /path/to/working/directory:/mnt /path/to/monai/container/monaicore150.sif python3 /mnt/train.py --flags...- singularity exec: Run a program inside the specified container.
- --nv: Enable NVIDIA GPU support. Omit on CPU-only systems.
- --bind /path/to/working/directory:/mnt: Mount host directory into container for data/scripts I/O.
- /path/to/monai/container/monaicore150.sif: Path to MONAI Singularity image.
- python3 /mnt/train.py ...: Command executed inside the container with relevant flags.
!!! Before running any script, update the bind mount and container path to your actual environment.
We provide a shell script that builds the MONAI-based container:
./build_container_v150.sh
Output: a folder named monaicore150 in your desired directory.
If your data already matches the following structure, you can skip preprocessing:
data/
imagesTr/ 111111.nii, 111112.nii, ...
imagesTs/ 111222.nii, 111223.nii, ...
labelsTr/ 111111.nii, 111112.nii, ...
labelsTs/ 111222.nii, 111223.nii, ...
dataset.json
Otherwise, ensure your raw data follows the Required Data Structure above, then run preprocessing.
Preprocessing Script Arguments:
- --data: Path to the main DOMINO data directory (required)
- --source-folders: Space-separated list of folder names within the data directory (required)
- --verbose: Enable verbose logging (optional)
Run:
./preprocess.sh
Output: nnU-Net style folders and dataset.json as shown above.
Training Script Arguments:
- --num_gpu: number of GPUs for training (default 3)
- --spatial_size: patch size (H, W, D) (default 64)
- --a_min_value: min pixel intensity for normalization (default 0)
- --N_classes: number of tissue classes (default 12)
- --a_max_value: max pixel intensity for normalization (default 255)
- --max_iteration: total training iterations (default 25000)
- --batch_size_train: training batch size (default 10)
- --model_save_name: filename prefix for saved model (default "unetr_v5_cos")
- --batch_size_validation: validation batch size (default 10)
- --json_name: dataset JSON filename (default "dataset.json")
- --data_dir: dataset directory (default "/mnt/training_pairs_v5/")
- --num_samples: number of data samples (default 24)
- --csv_matrixpenalty: csv file containing a matrix of penalty values (default /mnt/hccm.csv)
Run:
./train.sh
Estimated time: ~1 hour for 100 iterations; ~24 hours for 25,000 iterations.
Outputs (saved under --data_dir, prefixed by --model_save_name, e.g., "DOMINO"):
- [model_save_name].pth: Trained PyTorch model
- [model_save_name]_Loss.csv: Training loss per evaluation interval
- [model_save_name]_training_metrics.pdf: Plots of training loss and validation mean Dice over iterations
- [model_save_name]_ValidationDice.csv: Validation mean Dice per evaluation interval
Testing Script Arguments:
- --num_gpu: number of GPUs (default 1)
- --spatial_size: sliding window patch size (default 256)
- --a_min_value: min pixel intensity (default 0)
- --N_classes: number of tissue classes (default 12)
- --a_max_value: max pixel intensity (default 255)
- --batch_size_test: test batch size (default 1)
- --model_load_name: model to load (default "unetr_v5_bfc.pth")
- --json_name: dataset JSON filename (default "dataset.json")
- --data_dir: dataset directory (default "/path/to/data/")
Run:
./test.sh
Note: Provide multiple GPUs to testing DOMINO, if you trained DOMINO using multiple GPUs. Outputs: segmentation maps for each test image saved as .nii.gz under:
[data_dir]/TestResults/[model_name]/
Each output preserves header and affine from the input image.
You can run preprocessing, training, or testing via Docker by uncommenting the desired command in docker-compose.yml and running:
docker compose up --build
After completion:
docker compose down
Published Docker image:
- Preprocess:
docker run -v "$(pwd)/data:/data" nikmk26/domino:latest preprocess --source-folders d1 d2 d3 --verbose
- Train:
docker run -v "$(pwd)/data:/data" nikmk26/domino:latest train --data_dir /data --model_save_name DOMINO --batch_size_train 1 --batch_size_val 1 --max_iteration 1000 --spatial_size 64 --json_name dataset.json --num_gpu 2 --num_samples 25
- Test:
docker run -v "$(pwd)/data:/data" nikmk26/domino:latest test --data_dir /data --model_load_name DOMINO.pth --spatial_size 32 --json_name dataset.json --num_gpu 1
You can use our pre-trained models for testing. Please fill out the request form before accessing DOMINO models. Download pre-trained models here
Reproducible capsule:
DOMINO CLI processes NIfTI (.nii or .nii.gz) files using the DOMINO model, with batch support. Full usage and examples are in the repo.
Prerequisites:
- Python 3.9+
- Ability to create virtual environments (python3-venv)
- Docker (optional)
Whole Head Segmentator is a web based tool which will let you test our models like GRACE, DOMINO and DOMINO++ directly on web which any downloads or extra steps, just upload and select which model you are willing to test. It also facilitates the viewing of MRI images in 2D and 3D. Disclaimer: Uploaded volumes are sent to the backend to compute segmentations. Results are not intended for direct clinical decision-making. Do not upload data with direct identifiers or data that violates your data use or ethics agreements.
If you use this code, please cite:
@InProceedings{stolte2022DOMINO,
author="Stolte, Skylar E. and Volle, Kyle and Indahlastari, Aprinda and Albizu, Alejandro and Woods, Adam J. and Brink, Kevin and Hale, Matthew and Fang, Ruogu",
title="DOMINO: Domain-aware Model Calibration in Medical Image Segmentation",
booktitle="Medical Image Computing and Computer Assisted Intervention (MICCAI) 2022",
year="2022",
url="https://arxiv.org/abs/2209.06077"
}
Supported by NIH/NIA (RF1AG071469, R01AG054077), NSF (1908299), and the NSF-AFRL INTERN Supplement (2130885). We acknowledge the NVIDIA AI Technology Center (NVAITC) for their suggestions, and thank Jiaqing Zhang for formatting assistance. Base model: UNETR (MONAI) — https://github.com/Project-MONAI/research-contributions/tree/main/UNETR
Discussion, suggestions, and questions:
Smart Medical Informatics Learning & Evaluation Laboratory, Dept. of Biomedical Engineering, University of Florida